As open source, big data tools enter the early stages of maturation, data engineers and data scientists will have many opportunities to use them to “work on stuff that matters”. Along those lines, computational biology and medicine are areas where skilled data professionals are already beginning to make an impact. I recently came across a compelling open source project from UC Berkeley’s AMPLab: ADAM is a processing engine and set of formats for genomics data.
Second-generation sequencing machines produce more detailed and thus much larger files for analysis (250+ GB file for each person). Existing data formats and tools are optimized for single-server processing and do not easily scale out. ADAM uses distributed computing tools and techniques to speedup key stages of the variant processing pipeline (including sorting and deduping):
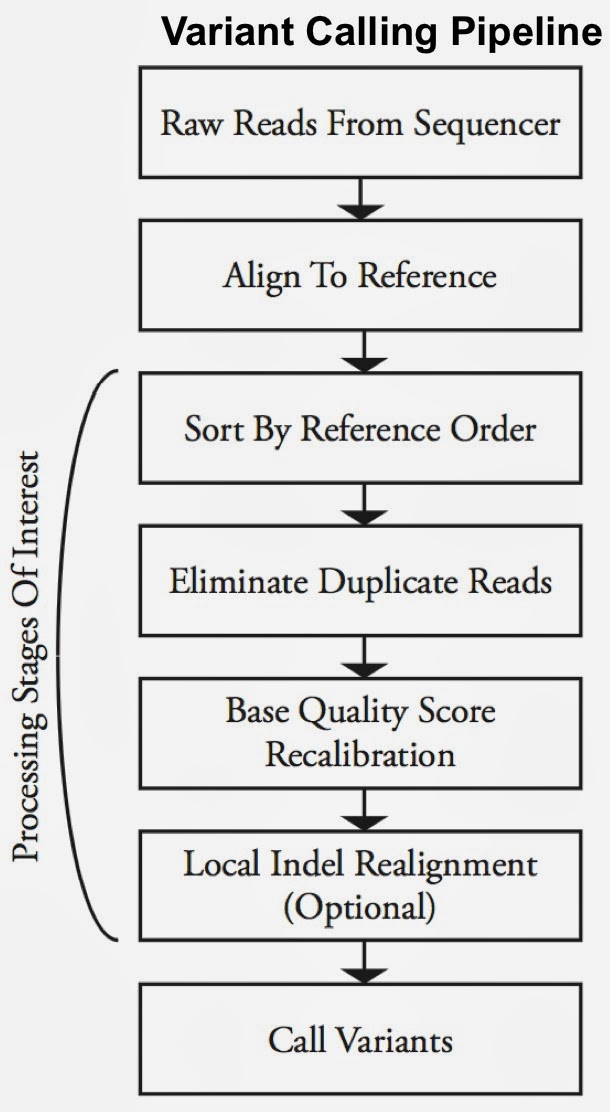
Very early on the designers of ADAM realized that a well-designed data schema (that specifies the representation of data when it is accessed) was key to having a system that could leverage existing big data tools. The ADAM format uses the Apache Avro data serialization system and comes with a human-readable schema that can be accessed using many programming languages (including C/C++/C#, Java/Scala, php, Python, Ruby). ADAM also includes a data format/access API implemented on top of Apache Avro and Parquet, and a data transformation API implemented on top of Apache Spark. Because it’s built with widely adopted tools, ADAM users can leverage components of the Hadoop (Impala, Hive, MapReduce) and BDAS (Shark, Spark, GraphX, MLbase) stacks for interactive and advanced analytics.
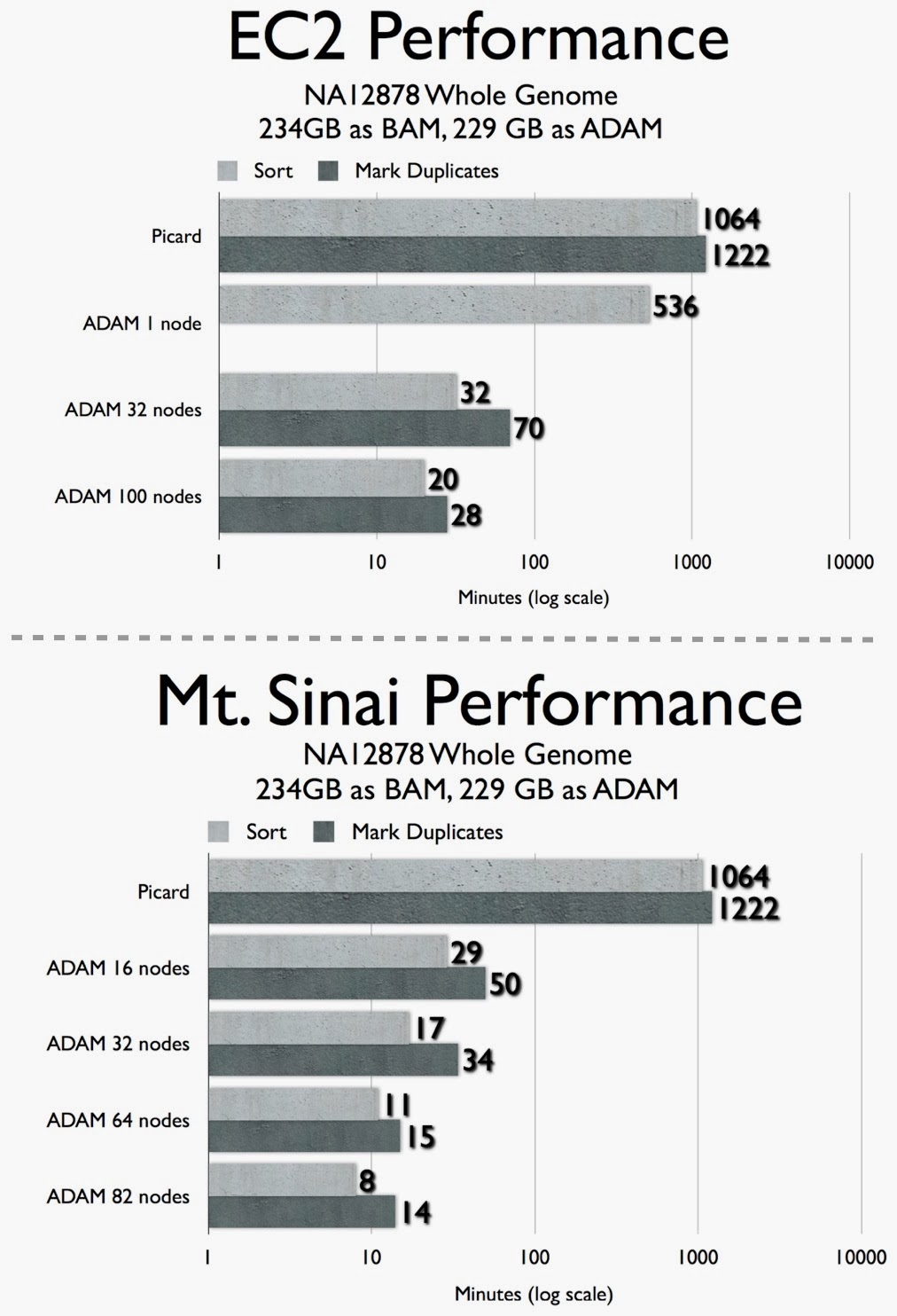
Although active development only started in Sept/2013, early results indicate that distributed computing tools and techniques lead to substantial speedups. Below are two recent tests from different computing clusters: : Amazon EC2 and a cluster at the Icahn School of Medicine at Mt. Sinai1. The combination of sorting and deduping took 38 hours using existing tools, but runs on less than an hour on a 32-node ADAM cluster.
Computational results like the ones above are drawing the attention of the science community: the AMPLab recently joined an Oregon Health & Science University (OHSU) research initiative to BeatAML (acute myeloid leukemia).
How to help
ADAM is a new project with a small codebase (11,000 lines of code). If you’re a big data hacker looking for a high-impact project to work on, consider contributing to the development of ADAM. Components are developed under an Apache License, so your contributions benefit the open source community. For details on how to contribute contact Matt Massie, lead developer of ADAM.
(0) This post is based on an extended conversation with Matt Massie. For more on ADAM, see this recent technical report.
(1) The cancer research program at the Icahn School of Medicine at Mt. Sinai, was the subject of a moving feature on Esquire magazine.
 O’Reilly Strata Conference — Strata brings together the leading minds in data science and big data — decision makers and practitioners driving the future of their businesses and technologies. Get the skills, tools, and strategies you need to make data work.
O’Reilly Strata Conference — Strata brings together the leading minds in data science and big data — decision makers and practitioners driving the future of their businesses and technologies. Get the skills, tools, and strategies you need to make data work.
Strata in Santa Clara: February 11-13 | Santa Clara, CA
